Using LLMs to Interface with FHIR Healthcare Database
Summary
Say you have a dataset that you want a large language model (LLM) to have access to so that it can answer questions relating to that dataset. One option would be to fine-tune the model itself by retraining on the new dataset, but there are a lot of problems with this approach. Retraining models is expensive and new parameter tuning can overwrite old information the model was initially trained on. Additionally, LLMs can hallucinate and make up information about the dataset. Finally, if the dataset contains sensitive information, then you likely won’t want to use it for retraining, as the LLM could give away that information to users without proper access. A better way to approach this problem is to use Retrieval-Augmented Generation (RAG) to connect the LLM to the dataset. This does not require any retraining of the model, and access to the data can be controlled at a user level. With this approach, the LLM can access the stored data and use it to answer questions. This post will walk through an in-depth example of utilizing an LLM with a graph database. Specifically, we will work with a FHIR database, and we will describe two separate approaches for querying this type of database.
Datasets/Model
Database
For this example, we will use a synthetic Fast Healthcare Interoperability Resources (FHIR) connected graph. FHIR is a system used to store healthcare information, formatted as an interconnected graph of resources, such as patients, treatments, visits, providers, and more. Thus, it can easily be represented as a graph database, through a platform such as Neo4j. FHIR is a complicated schema, with over 25 different node types and hundreds of attribute and relationship types. Below is an example of a subgraph from this type of database, representing the nodes that are all connected to just one single patient node.
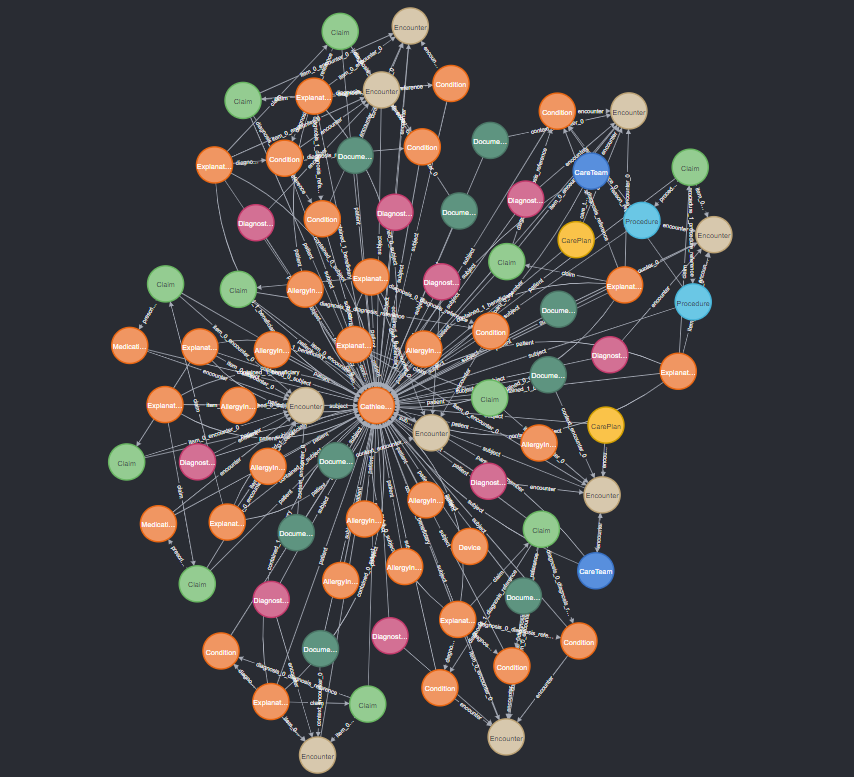
Even just a subgraph of one patient’s neighbors is complicated, and this will require some special techniques to successfully work with a database of this complexity. The main problem is simply connecting the LLM to the database so that it can access it to answer queries. Further in this post, I will describe two separate techniques for accomplishing this, which each have their own strengths and weaknesses.
More information about the data itself can be found here.
Vector Store Approach
This approach converts the entire graph database into a new format that the LLM can work with more easily. First, each node’s attributes are summarized into sentences to form a single paragraph that contains all information about that node. Then, these paragraphs are embedded using a vector embedding model, which converts each paragraph into a vector of numbers. These vectors are kept in a vector store database, for which there are multiple algorithms for efficient similarity searches. Using a similarity search, the node that is most relevant to an input query can be retrieved and used to answer a question. More details about this vector store method for storing graph databases can be found here.
Once the most relevant node is retrieved from the vector store, it can be used in a query on the original graph database to gather all connected nodes to that node. This generates a subgraph containing the relevant parts of the database, and the text summary used to generate the embeddings can be retrieved from each neighbor and included in the context provided to the LLM. This way, the LLM can answer questions related to multiple connected nodes, because it has access to the text summaries from those nodes.
However, due to the size and complexity of the schema, there are many times when the amount of information connected to the central node exceeds the LLM’s context size, meaning it is too much data for the LLM to use. We can limit this data by only retrieving nodes of types that are determined to be relevant to the query. In order to determine which node types would be relevant, we use another LLM. By passing in the user’s input query and a description of each node type and when they would be relevant, the LLM can return a simple list of node types relevant to that question that we can use in the graph query. Then, the query can be limited to only those node types, and the gathered context for the final response will be small enough for the LLM to use.
In the example below, if I ask the question “What is the heart rate of patient Adan632 Cassin499 on their most recent measurement?”, then the similarity search can find the patient node first, and then the relevant nodes connected to Adan632’s node would be retrieved as well, which will contain the patient’s vital sign measurements at every visit. The LLM can then use its natural text reasoning skills to determine the most recent measurement and provide it to the user.
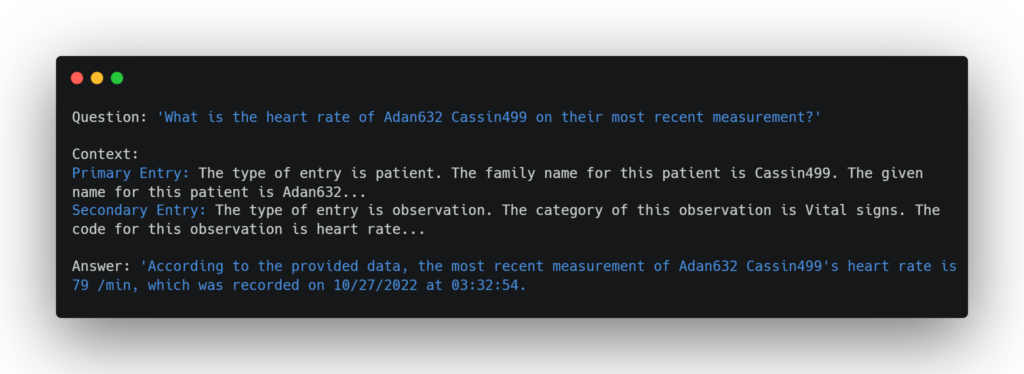
This method works very well for a variety of query types. It excels when asking questions about specific patients or conditions, and due to the similarity search method and the strength of vector embeddings, it is very flexible to rewordings and misspellings. However, there is a weakness with this method, as any queries involving a large number of nodes will not be answered correctly. For example, take the question “How many patients are under the age of 30 by the end of 2021?”. To answer this question, the LLM would need data from every patient to determine their ages, but since these patient nodes are unconnected or distantly connected, it won’t be able to retrieve all of them at once. Therefore, a different, more flexible method is required for these types of queries.
Direct Querying Approach
Newer developments in LLM capabilities now allow LLMs to directly query graph databases. LangChain provides a pipeline through which user questions are reworked into valid queries that can run on Neo4j hosted databases, using the Cypher query language. The results of the query are then returned to the LLM to be used in the answer.
Still, without any modifications, the Cypher queries that are generated are often incorrect, frequently misinterpreting the meanings of certain attributes. This is because many attributes in the FHIR schema are unclear in name alone. To address this, we can pass in additional context that provides examples for each node type, which can explain what each attribute actually stores. Similarly to in the first approach, we limit this to only node types that are relevant to the question, so that the context is not overwhelmed with irrelevant information. Providing these examples led to a significant improvement in the quality of the queries. This method is much more flexible than the first approach in the types of queries that it can respond to. For example, the question “How many patients are under the age of 30 by the end of 2021?” now returns the correct answer. The LLM is able to write a correct query that is run on the database to return the result.
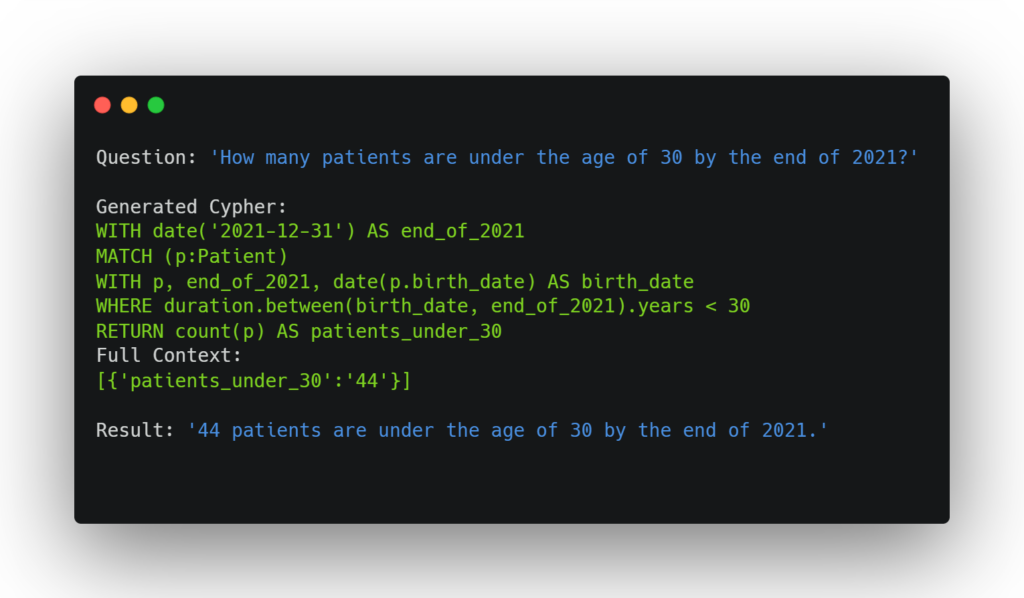
This method is much simpler than the first, requiring less setup and working more intuitively. However, a disadvantage is that it is less flexible to differently-worded queries compared to the first approach. For example, asking for the ‘body weight’ of a patient at a certain observation will work because the measurement’s name is ‘body weight’, but just asking for the ‘weight’ of a patient may not match if the LLM does not correctly match your terminology with that of the database, because the query will try to find an attribute named ‘weight’ when none exists.
Access
Here, example code will be given to perform the direct querying approach. The vector store approach is more complicated to set up and code, but examples can be found here.
First, we must import the necessary packages and set up two fundamental parts: the LLM and the graph database. A variety of LLM models can be used here, but the example given here uses GPT 4-o. Additionally, the graph DB is hosted locally using Neo4j.
from langchain_core.prompts import PromptTemplate
from langchain_openai import ChatOpenAI
from langchain.chains import create_retrieval_chain
from langchain.chains import GraphCypherQAChain
from langchain_community.graphs import Neo4jGraph
import json
from neo4j import GraphDatabase
with open('config.json') as user_file:
config = json.load(user_file)
llm = ChatOpenAI(
model_name="gpt-4o",
openai_api_key=config["openai_api_key"],
temperature=0
)
driver = GraphDatabase.driver(uri="bolt://localhost:7687", auth = (config["username"], config["password"]))
def get_database_localhost():
NEO4J_URI = "bolt://localhost:7687"
NEO4J_USERNAME = config["username"]
NEO4J_PASSWORD = config["password"]
graph = Neo4jGraph(url=NEO4J_URI, username=NEO4J_USERNAME, password=NEO4J_PASSWORD)
return graph
With these defined, it is only a few lines of code to define a query and pass it to the LLM along with the graph database object. Additionally, we need to define a prompt for the LLM. In the most simple case, this can be done like so.
graph = get_database_localhost()
question = "How many patients are under the age of 30 by the end of 2021?"
cypher_prompt = """
You are an assistant that generates valid Cypher queries based on the provided schema and examples.
Use only the provided relationship types and properties in the schema.
Do not use any other relationship types or properties that are not provided.
Do not write any queries to create, edit, or delete nodes. You have read-only access to this database.
Important: Return only the Cypher query, with no additional text or notes.
Generate a Cypher query to answer the following question: {question}
"""
prompt_cypher = PromptTemplate(template=cypher_prompt, input_variables=['question'])
chain = GraphCypherQAChain.from_llm(llm, graph=graph, verbose=True, cypher_prompt=prompt_cypher)
input = {"query":question}
print(chain.invoke(input))This is all the necessary code to interface with a graph database using an LLM, and it will work well for simple cases. Langchain will automatically gather the schema for the database and use it as context for the LLM so that you do not need to manually pass it in. Thus, it is very simple to set this up and begin querying. However, as described in the “Direct Querying Approach” section, more steps are sometimes needed for complicated schemas such as FHIR. For example, the schema that Langchain automatically gathers will include all node, attribute, and relationship types, but it won’t provide much detail. The graph schema does not include information about what each attribute actually means, and will provide no examples of the correct formatting and data types that may be needed for proper querying. Thus, if the schema is complex or the naming is unintuitive, the LLM may not be able to decipher which attributes and relationships it needs to use to answer a question. To combat this, we can provide example nodes for relevant node types that provide this information, allowing the LLM to properly understand the database. To do this, we can first write a simple LLM call to determine what node types are relevant to the question.
def determine_relevant_nodes(question):
template = """
Task: The user input will concern a question related to a graph database, which contains many node types. Your job is to determine what node types are relevant
to this particular user question. Your response should be a comma separated list of node types, if there are more than one, with no additional text.
Here are the node types and when they are relevant:
Claim: related to billing, insurance, and pricing for procedures and encounters
Condition: relevant for conditions and diagnoses
Encounter: related to specific patient visits and what a patient was there for
Observation: relevant for a particular measurement of a patient at a given time, such as vital signs or BMI
Patient: relevant for questions relating to specific patients, including demographic info such as locations, DOBs, and other statuses
Question: {question}"""
prompt = PromptTemplate.from_template(template)
llm_chain = prompt | llm
response = llm_chain.invoke({'question': question})
return response.contentThese are just a few examples of node types and do not contain every FHIR node type. However, we have found that a question of this format works very well at gathering relevant node types. Once we have a list of node types, we can use that to gather example node data.
def get_schema_for_node_types(node_labels):
all_info = ""
with driver.session() as session:
for label in node_labels:
query = f"""MATCH (n:{label})
WITH n
LIMIT 1 RETURN n"""
result = session.run(query)
results = [record.data() for record in result]
info = label+": "+str(results[0]).replace('{', '(').replace('}', ')')
relationships = str(session.read_transaction(get_relationship_types, label))
all_info = all_info + info + ' Relationship types: '+relationships+ '\n'
return all_info
def get_relationship_types(tx, node_label):
query = f"""
MATCH (n:{node_label})-[r]-()
RETURN DISTINCT type(r) AS relationship
"""
result = tx.run(query)
return [record['relationship'] for record in result]The get_schema_for_node_types function creates a textual representation of an example node for each type, listing each attribute and their values, as well as the relationship types connected to that example node. This string can then be included as part of the prompt, giving extra information to the LLM that allows it to complete its task more effectively. With these relatively simple adjustments, the accuracy and performance of the LLM can be greatly improved, allowing for efficient and effective querying of graph databases.
If you’re interested in other kinds of work related to LLM interfacing and LangChain, check out our GitHub repo.
Ownership
This was an internal experimental project to explore the available techniques that could be used to query graph databases using LLMs.
This topic was a part of our LLM Workshop Presentation on 09/18/2024. You can watch that presentation here.
Resources
Aaron Mullen was the lead developer on this project.

