In 2023, Kentucky had the fifth-largest drug overdose fatality rate in the United States, and 79% of those deaths involved opioids. To help combat the opioid epidemic in Kentucky, the CAAI works with the Rapid Actionable Data for Opioid Response in KY (RADOR-KY) team to provide support with machine learning techniques. Overall, the team works to create a statewide surveillance system to monitor and respond to the opioid crisis, collecting data from a variety of sources and agencies around Kentucky. One key part of the project is predictive analytics, using machine learning techniques to forecast future trends of opioid overdoses in different areas of Kentucky. The goal is to provide accurate forecasts based on different geographical levels to identify which areas of the state are likely to be the most “high risk” in future weeks or months. With this information, adequate support could be prepared and provided to those areas with the hope to treat victims in time and reduce the number of deaths associated with opioid-related incidents.

Data Pre-Processing
The first step was to analyze what geographical level would be most appropriate for building and training a forecasting model. The data we use to track suspected opioid overdose incidents comes from Kentucky Emergency Medical Services (EMS) responses, beginning in January 2017 and continually updating. With this data, we could group incidents based on six different geographical levels: state, county, zip code, tract, blockgroup, and block. Through experimentation, it seemed that the county level was likely the most appropriate scale. State level is too broad for useful results, while any level smaller than zip code proved to be too sparse. Smaller geographical levels contain too few positive examples of incidents for any model to successfully learn the trends of each area. However, data sparsity remains a problem even at the county level in less-populated areas, so we have also worked with Area Development Districts (ADDs), which are larger groupings of counties. Additionally, the temporal level was chosen to be at the monthly scale, rather than yearly or weekly, due to early testing results suggesting the best performance at monthly levels.
Additional Data
Time series forecasting models typically are able to use previous values of your target series to determine the trends and patterns that it uses to predict into the future. However, many models can also use additional data sources to aid in the predictions, called covariates. A variety of data from different sources has been tested as covariates, including:
- Temperature and precipitation monthly averages
- Social determinants of health for each county, such as unemployment rate, vehicle access, and age distributions
- Aggregated Medicaid claims containing counts of individuals diagnosed with or receiving treatment for Opioid Use Disorder, among other measurements
- Kentucky State Police drug seizures for opioid substances
- Kentucky Department of Corrections substance use risk measures for inmate intakes and releases
- Naloxone distribution counts for each county
Some covariates, such as weather, are future covariates, meaning that their values are known into the future and can be used at prediction time. Others, such as drug seizures, are past covariates, so their historical data can be used to capture trends and correlations, but future values are not known ahead of time. The social determinants of health are a static covariate, meaning the value does not change over time, but it can still be used to distinguish between the characteristics of different areas.
Models and Training
Several different models have been evaluated for forecasting. First, the Histogram-based Gradient Boosting Regressor is the simplest and most general model, using both statistical and machine learning methods. This models works by building a large ensemble of decision trees that use past values and covariates to create forecasts. Second, the N-Linear model is a simple, one-layer neural network built specifically for time series forecasting. Finally, the Temporal Fusion Transformer (TFT) is a large deep learning architecture, also built for forecasting. This is the most complex model evaluated here, using multiple layers and a self-attention mechanism to weight the importance of data at different time steps. These models were chosen among many others for two primary reasons. First, they are able to support all types of covariates, including future, past, and static. Many other models, particularly statistical methods, can only utilize future covariates when making predictions. Second, these models possess multivariate capabilities, allowing for separate series, such as each county or ADD, to be trained and evaluated with one model. That way, the model can learn the overall trends that each region shares, while also picking up on exclusive patterns specific to each region.
Before training, pre-processing techniques were performed to normalize each grouping’s data to its own mean and address missing values. Normalization is required because many of the regions have vastly different scales when counting opioid overdoses, and normalizing these ensures that the model considers all regions equally. Addressing missing values is required because many of the data sources for the covariates have misaligned data availability time frames. These missing values were filled with a constant value, and for each variable, an additional binary flag was created to indicate whether the data was originally missing.
When training, we have tested a variety of configurations of models and covariates to determine the most effective methods for creating accurate predictions. To do this, each model is evaluated separately, as well as each covariate. Different combinations of covariates are also tested, but generally, including more than only a couple of covariates can overwhelm the model and lower performance.
Results
Each run was evaluated with the Root Mean Squared Error (RMSE), which is a common performance metric for time series forecasting. Because it is a measure of prediction error, a lower value is better.
The N-Linear model outperformed the other two models in most cases, achieving a lowest RMSE of 0.1158, compared to 0.1272 for Gradient Boosting and 0.1462 for the TFT. The N-Linear model likely performs the best because it strikes the best balance between generalizability and complexity. The gradient boosting model works as a good baseline as an easy, general model, and the TFT proves to likely be too complex for this use case.
The most effective covariates across models were found to be the drug seizures and Medicaid claim data, achieving lower RMSEs across all models when compared to other covariates. The biggest impact on error, though, was the choice of geographical grouping. Every model performed significantly better when ADD groupings were used instead of counties. The average best RMSE across models for the ADD groupings was 0.1297, while the average for the counties was 0.1830. This is likely due to the sparsity of data in many areas at the county level, as the ADD aggregations increase the number of overdose events for each series. This leads to lower error because the trends are more easily deciphered at larger scales; changes in counts at a small scale are more influenced by randomness.
Example results are shown below for two separate ADDs: KIPDA (A) and Lake Cumberland (B). These figures highlight how the model can learn the trends that all groupings have in common, while adjusting for each individual time series. Both forecasts show a decrease of overdoses at the beginning of 2024 and an increase again afterwards, but the exact shape and position of the forecasts differs.
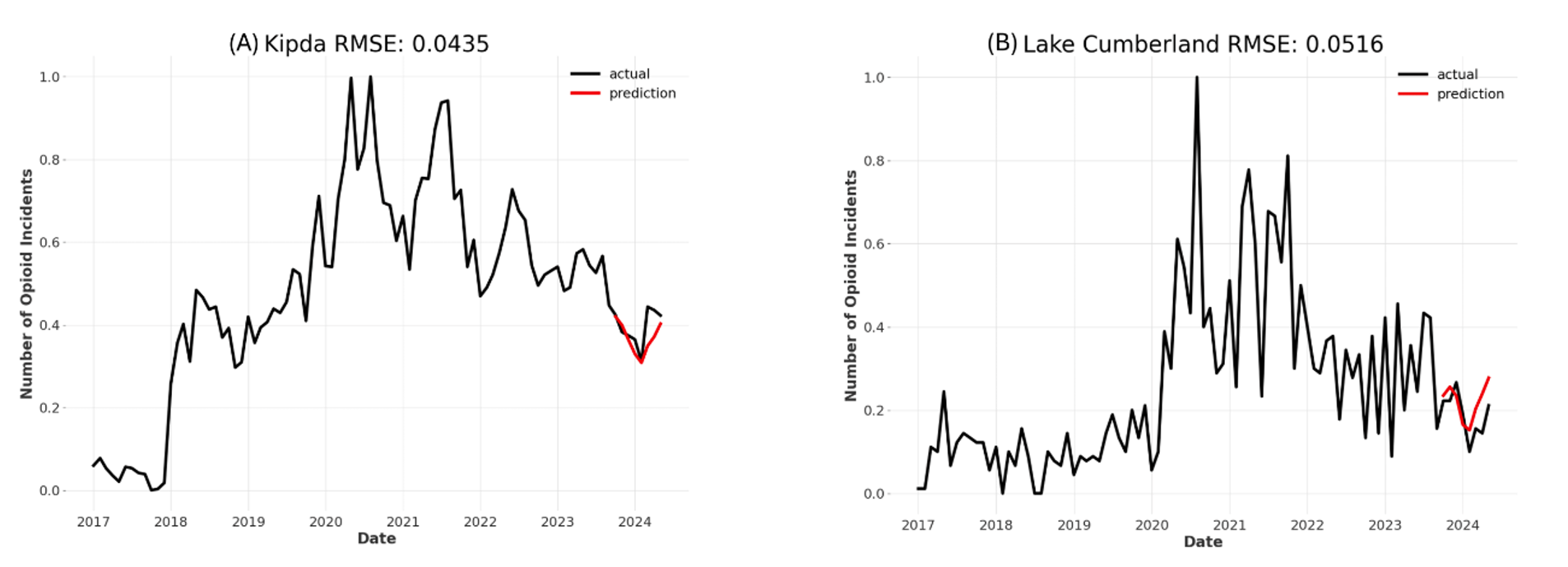
We will continue to test new models and external data sources to improve forecasting accuracy. Still, the current results show that forecasting opioid overdoses around Kentucky is possible with limited error and will prove useful to state agencies for determining when and where opioid overdoses can be expected to increase or decrease.
Resources
A paper detailing this topic was submitted to the American Medical Informatics Association in 2024 and can be found here.
A presentation covering this topic was given at UK’s AI/ML seminar on 10/17/2024. The slides for that presentation can be found here.

