Center for Applied Artificial Intelligence
Where cutting edge technology meets
translational science.
The Center for Applied Artificial Intelligence functions within the University of Kentucky Institute for Biomedical Informatics to explore new technologies, foster innovation, and support the application of artificial intelligence in translational science.

Recent Projects
Interact with our state of the art Large Language Model in a convenient browser environment.
Compare a vanilla Large Language Model's performance against one that has been given a trained adapter.
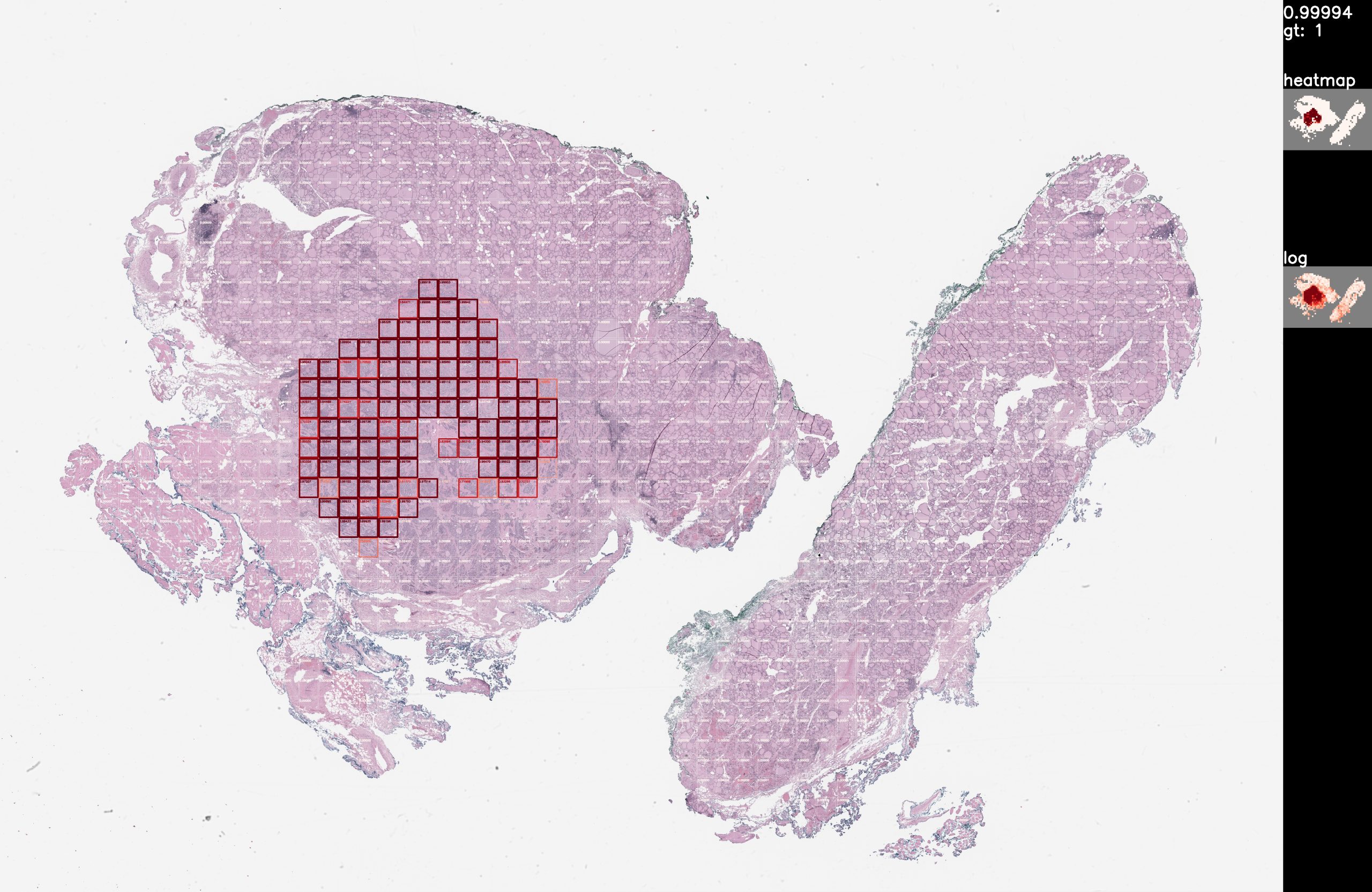
To avoid unnecessary surgical removal of lymph nodes in thyroid cancer patients, we would like to predict from thyroid whole slide images whether the tumor is aggressive or not. As a preparation to this goal, we would also like to predict the presence and location of the tumor in thyroid whole slide images. Given a whole slide image of a thyroid tissue, detect the tumor regions. For example, this is a visualization of a true positive detection from the validation dataset: Train/test at x5 magnification, rescaled from x40 487 train slides (80%) 123 validation slides (20%) 164 lymph node slides, visualization only 577,668 total tiles @ 224×224 ~18 hours training on RTX 5000 Github (private): https://ipopgit.uky.edu/ibi-pathology/mskcc-mil-nature-medicine-2019 Contact authors for model
Hepatitis A Predictor We are working on hepatitis A prediction modeling. Details will be limited until the associated paper is published. Details provided on publication Details provided on publication https://medicine.ai.uky.edu/api/hepaa Github (private): https://github.com/codybum/hepa_a_pred Contact authors for model
COVID-19 Cell Detector Here are our results in a COVID-19 challenge hosted by telesto.ai. In this challenge the aim is to build a tool which is capable to diagnose COVID-19 (healthy or infected) from microscopic images of blood tests. The challenge provides a set of training images with labels and a training set for prediction. Results of image prediction are submitted to the challenge host. We are able to achieve high (+97%) precision, recall, auc, and acc predictions on hold-out images from the training set. On the training set we achieved 92.13% acc (only reported measure), tying for first place as of 8/15/2020. Details on the data set are provided by the challenge host: https://telesto.ai/competition/covid-19/dataset We are provided +433 images with labels (healthy or infected) for training and 217 images without labels for testing. The training set shows signs of curation, as despite various attempts the testing prediction results are 7-10% worse than hold-out testing sets of training images. Github (private): https://github.com/codybum/covid19-ai Contact authors for model
Soft Tissue B/M We are attempting to classify benign vs malignant in soft tissue slides. The training accuracy seems to improve, especially with the 1000 tiles data set. However, validation is another story. It is clear that nothing useful is being learned by the models. We have a limited number (~200) slides to work with on this project and unfortunately very few of the slides are the same body part. I am told that most soft tissue slides are mostly tumor, so B/M methods for many organ and tissue types that look for tumors don’t yield good results. The only positive here is that in attempts to generate something better than a coin flip, we spent the time to develop model code that would run across and (almost) full utilize many GPUs.
The computers do not know tissue from glass. Methods must be used to identify image regions (tiles) that contain tissue. Our efforts makes use of the excellent work: https://github.com/deroneriksson/python-wsi-preprocessing Process: -Apply filters for tissue segmentation -Mask out non-tissue by setting non-tissue pixels to 0 for their red, green, and blue channels. -This algorithm includes a green channel mask, a grays mask, a red pen mask, a green pen mask, and a blue pen mask. -Threshold mechanisms: basic, hysteresis, and Otsu
We have been working with BERT[1], a natural language processing (NLP) AI model Google released a few years ago. BERT can be used for a number of NLP tasks, including multi-label classification. Using gross pathology reports, we have training our own language models using BERT to be used in the fine-tuning of pathology-focused NLP models. The graphs show perplexity and loss scores for 10, 15, and 25 epochs.
We have been working with BERT[1], a natural language processing (NLP) AI model Google released a few years ago. BERT can be used for a number of NLP tasks, including multi-label classification. I have been using BERT to determine the tissue type (part of determining case complexity for scheduling) from the gross pathology report, and I am getting good results. We are able to achieve 95% accuracy on 38 labels using just 200 reports per label. People are starting to augment (training with unclassified data) BERT models with clinical text[2], we have done with surgical pathology reports, which in my case seems to improve results 2-5%. It would be reasonable for us to train models on specific types of reports from other areas (radiology, neurology, etc.), which might further improve accuracy. This is not fine-tuning training, but rather changing the structure transformer model to better work with the domain vocabulary. Hypothetically (and my limited experience), a Pathology-BERT performs better on Pathology fine-tuned models, than a base BERT model. [1] https://en.wikipedia.org/wiki/BERT_(language_model) [2] https://www.aclweb.org/anthology/W19-1909.pdf